Investigation of cell surface changes due to detergent treatment using scanning ion conductance microscopy (SICM)
Research Application Technology Center, Park Systems Corp.
Abstract
In biochemical processing, detergents play an important role in modification of the cell surface membrane. Detergents are widely used to disrupt the cell membrane for the release of soluble inter- and intracellular molecules and cause significant implications in the histological study of cells. Although many studies report on the influence of detergent processing on the membrane permeability, the structural effects of the cell membrane at the nano level are not well understood. In this study, Scanning Ion Conductance Microscopy (SICM) is performed to compare structural changes of the Huh7 cell surface with and without treatment of varying concentrations of the non-ionic detergent Triton X-100. By using the distance dependence of an ionic current as feedback parameter, SICM can non-invasively image the increasing surface deterioration of the cell with increasing detergent concentration. Because of this capability, the images taken using SICM offer new insights on the interactions of cells and detergents.
Introduction
Detergents are crucial and fundamental chemical agents in biological study, with particular emphasis in cell biology [1], [2]. Detergent molecules contain both hydrophobic and hydrophilic chemical groups, which easily interact with cellular membranes. Many studies have been published regarding the role of detergent on the modification of a cell membrane; the primary role of detergent is disruption of cell membranes and formation of tiny holes in the membrane surface. Through these holes, internal cell membrane proteins can be extracted, while external antibodies can be introduced. Based on their charge properties, detergents are classified as non-ionic, ionic and zwitterionic [3], [4], [5]. Among these, the non-ionic detergent, Triton X-100, has a rigid and bulky non-polar head group. Thereby it can bind to water-insoluble, hydrophobic compounds, such as cell membrane proteins and lipids, and solubilize them in aqueous media. The detergent’s interaction with cell membrane components effectively changes the membrane integrity by creating holes or even leading to a cell surface collapse [6].
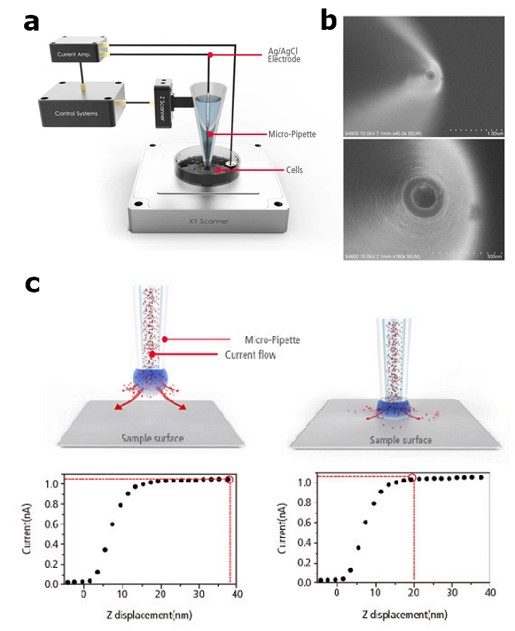
Figure 1. Basic set-up and principle of SICM. Schematic view of simple set-up of SICM (a),
SEM image of the nano-pipette (b) Schematic diagrams and corresponding ion current responses
of the SICM principle (c).
To image at nanoscale the cell surface changes due to detergent treatment, Scanning Ion Conductance Microscopy (SICM) is ideally suited. It is a zero force, non-contact scanning probe microscopy (SPM) technique as it uses the ion current feedback to scan and image the sample. In SICM, a nano-pipette scans the sample surface in solution, while an ionic current flow between an electrode within the pipette (pipette electrode) and an electrode in the sample solution (bath electrode) is detected [7]. The detection of the ion current provides a feedback signal that allows maintaining a constant tip-sample distance (Figure 1). Therefore, SICM can obtain topographical information without physical contact with the sample surface, which makes this technique truly non-invasive. The lack of surface contact associated with the SICM technique has the added benefit of data acquisition without damaging the cellular surface [8].
In this note, Huh7 cells surface morphology changes due to the treatment of the non-ionic detergent Triton X-100 were investigated using SICM. Triton X-100 was utilized at varying concentrations (from 0.01–1%). The nano-scale membrane structure of non-ionic detergent treated cell membrane was monitored and structural changes of cell surface depended on detergent concentration were obtained.
Experimental set-up
Cell sample
The Huh7 cells (ATCC, USA) were cultured in Dulbecco’s modified eagle medium (DMEM; Invitrogen Life Technique, USA) and complemented with 10% fetal bovine serum (Thermo Fisher Scientific, USA) together with 1% penicillin/streptomycin (Invitrogen Life Technique, USA) at 37°C in a humidified atmosphere with a concentration of 5% CO2. Cells were harvested after trypsinization, washed and re-suspended at a concentration of 1×105cells/mL in normal growth medium for subculture. A 1mL aliquot of the cell containing medium was seeded on the experimental dish. The cell concentration was adjusted to 1×104/mL on the 35mm dish.
Scanning ion conductance microscopy set-up
The set-up for cell imaging consisted of the Park Systems SPM system (Park NX12, Park Systems, Korea) equipped with an inverted optical microscope (Nikon Corp., Japan), specifically designed for biological applications. The inner diameter of nano-pipette was 80-100 nm. The reference ion current was 1 nA with applied 0.1 V of bias. The general set-point for SICM imaging was 2% decreased value from the reference ion current and averaged imaging time was approximately 25 minutes.
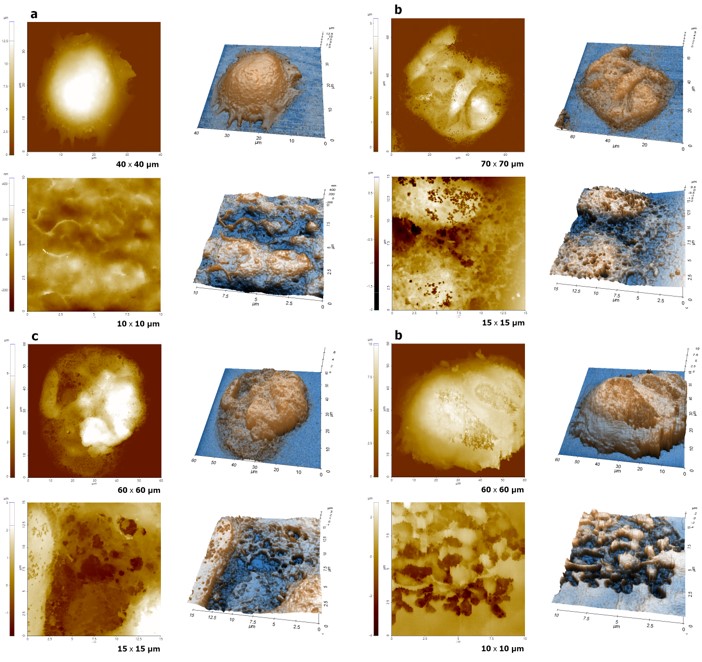
Figure 2. Cell surface images obtained by SICM: Untreated cell (a); cell treated with 0.01% Triton X-100 (b);
cell treated with 0.1% Triton X-100 (c) and a cell treated with 1% Triton X-100 treated cell (d).
All SICM images were performed with approximately 80 nm of inner diameter pipette in liquid.
Results and Discussion
To explore the detergent-treated cell surface, the Approach-Retract-Scan (ARS) mode (also known as hopping mode) of Scanning Ion Conductance Microscopy (SICM) was performed on cells treated with 0.01%, 0.1% and 1% of Triton X-100 as well as an untreated cell. Figure 2 shows topographical information of both untreated (Fig. 2a) and treated cell surfaces (Fig. 2b, 2c and 2d). As shown in figure 2, there are significant differences between non treated and detergent treated cell surface, even at a small detergent concentration (0.01%, Fig. 2b). Smooth and clean structures were observed on the untreated cell surface with a root mean square roughness (RMS) of 72 nm; however, many tiny holes and disruptive surface structures were seen on the detergent-treated cell membrane. The RMS values of 0.01%, 0.1% and 1% are 407 nm, 950 nm and 1,058 nm, respectively. The changes of the cell surface including the increased roughness are due to the interaction of the cell membrane with detergent molecules and resulted in cell surface fractures. Because of these changes, proteins, peptides and lipids can be extracted from or penetrated into a cell. Also, depending on detergent concentration, different disruptive levels are observed on the cell surface. Cells treated with higher concentrations of detergent have bigger holes and a more fractured surface.
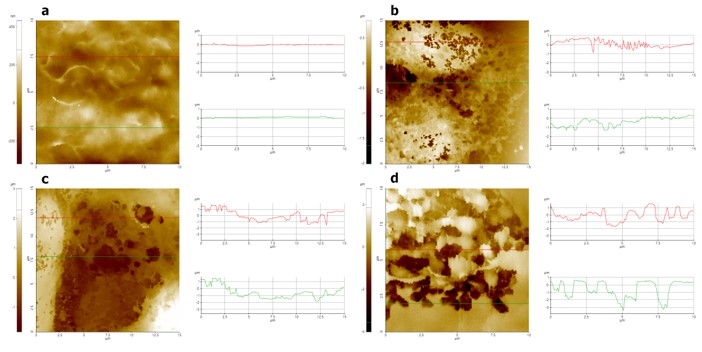
Figure 3. Line profile analyses of untreated and detergent-treated cell surfaces: Non treated cell (a); 0.01% Triton X-100 treated cell (b); 0.1% Triton X-100 treated cell (c) and 1% Triton X-100 treated cell (d).
The line profile analysis shown in Figure 3 clearly illustrates this phenomenon. For the untreated cell, no convex or concave lines were observed on the cellular surface. However, for cells treated with even low doses of detergent, many small holes and irregular line profiles were clearly visible. The depth and width of the hole structure were approximately 0.6 μm and 0.2 μm, respectively, for the 0.01% Triton X-100 treated cell. On the other hand, at higher concentrations of cells treated with Triton X-100 (0.1 and 1%), hole structures with depth and width of approximately 1~2 µm were estimated. This suggests that increasing the detergent concentration increased the interaction of the detergent with the cell membrane as well as increased changes to the cellular surface.
Conclusions
This study compared the morphological differences between cell membranes treated with varying concentrations of Triton X-100 detergent and untreated cells. Scanning ion conductance microscopy (SICM) measurements indicated significant surface changes to cell membranes when they were exposed to Triton X-100 concentrations (which varied from 0.01% to 1%). These findings provide understanding of how a detergent reacts with a cell membrane and opens the door for further examination of substrate transport in or out of a cellular component using a detergent treatment as one of the steps in that process. As one of the scanning probe microscopy techniques, SICM enables you to monitor real features of a cell surface using nano-pipette with ion current feedback, without any force sensing. Utilizing this technique, high resolution images can be obtained without inflicting damage to the cell surface and it is poised to serve as a promising tool in biological applications.
Reference
1. Oliver, C., &Jamur, M. C. (Eds.). (2010). Immunocytochemical methods and protocols. Humana Press.
2. Jamur, M. C., & Oliver, C. (2010). Permeabilization of cell membranes. In Immunocytochemical methods and protocols(pp. 63-66). Humana Press.
3. Otway, S., & Robinson, D. S. (1967). The use of a non‐ionic detergent (Triton WR 1339) to determine rates of triglyceride entry into the circulation of the rat under different physiological conditions. The Journal of Physiology, 190(2), 321-332.
4. Filip, C., Fletcher, G., Wulff, J. L., & Earhart, C. F. (1973). Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. Journal of bacteriology, 115(3), 717-722.
5. Perdew, G. H., Schaup, H. W., &Selivonchick, D. P. (1983). The use of a zwitterionic detergent in two-dimensional gel electrophoresis of trout liver microsomes. Analytical biochemistry, 135(2), 453-455.
6. Apgar, J. R., Herrmann, S. H., Robinson, J. M., &Mescher, M. F. (1985). Triton X-100 extraction of P815 tumor cells: evidence for a plasma membrane skeleton structure. The Journal of cell biology, 100(5), 1369-1378.
7. Hansma, P. K., Drake, B., Marti, O., Gould, S. A. C., & Prater, C. B. (1989). The scanning ion-conductance microscope. Science, 243(4891), 641-643.
8. Novak, P., Li, C., Shevchuk, A. I., Stepanyan, R., Caldwell, M., Hughes, S., & Moss, G. W. (2009). Nanoscale live-cell imaging using hopping probe ion conductance microscopy. Nature methods, 6(4), 279.
